Solving Crystal Structures From Two-wavelength X
advertisement
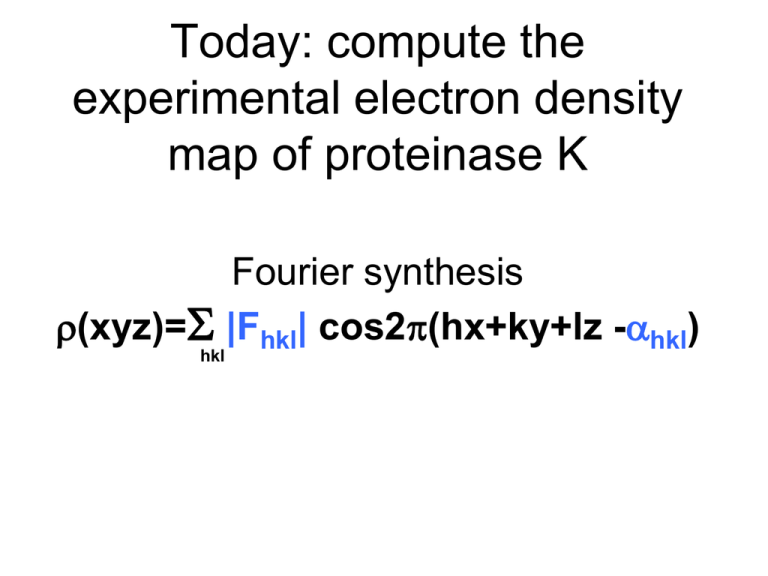
Today: compute the experimental electron density map of proteinase K Fourier synthesis r(xyz)=S |Fhkl| cos2p(hx+ky+lz -ahkl) hkl 3 Crystals 5 Measured Quantities Native H 1 1 1 1 1 1 1 1 1 K 1 1 1 1 1 1 1 1 2 L 1 2 3 4 5 6 7 8 0 PCMBS (Hg) |FP| |FPH (+)| |FPH (-)| 681.4 725.8 722.4 752.8 733.6 695.3 332.1 444.5 456.2 526.9 575.8 564.7 719.2 827.8 805.4 358.4 349.8 354.2 273.3 359.4 390.8 400.7 362.5 411.2 162.5 73.8 132.3 EuCl3 (Eu) |FPH (+)| |FPH (-)|) 730.8 707.6 813.9 805.3 296.1 312.5 527.4 518.3 759.6 766.3 375.6 358.9 300.5 286.6 396.7 411.5 149.8 159.8 Measureable differences 1) Isomorphous differences (between native and derivative) 2) Anomalous Differences (i.e. differences between Friedel pairs): 3) Dispersive differences (differences due to changing the wavelength): FP(hkl)=FPH (hkl) - fH(hkl) FP(hkl)=FPH(-h-k-l)* - fH(-h-k-l)* FP(hkl)=FPH (hkl) ln - fH(hkl) l n 3 Crystals 5 Measured Quantities Native H 1 1 1 1 1 1 1 1 1 K 1 1 1 1 1 1 1 1 2 L 1 2 3 4 5 6 7 8 0 PCMBS (Hg) |FP| |FPH (+)| |FPH (-)| 681.4 725.8 722.4 752.8 733.6 695.3 332.1 444.5 456.2 526.9 575.8 564.7 719.2 827.8 805.4 358.4 349.8 354.2 273.3 359.4 390.8 400.7 362.5 411.2 162.5 73.8 132.3 EuCl3 (Eu) |FPH (+)| |FPH (-)| 730.8 707.6 813.9 805.3 296.1 312.5 527.4 518.3 759.6 766.3 375.6 358.9 300.5 286.6 396.7 411.5 149.8 159.8 For EuCl3 (Eu) PCMBS (Hg) Vector equations for this experiment Isomorphous and Anomalous Differences Isomorphous and Anomalous Differences FP(hkl)=FPHg (hkl) - fHg(hkl) FP(hkl)=FPHg(-h-k-l)* - fHg(-h-k-l)* FP(hkl)=FPEu (hkl) - fEu(hkl) FP(hkl)=FPEu(-h-k-l)* - fEu(-h-k-l)* Vector equations for this experiment Isomorphous Differences FP(hkl)=FPHg (hkl) - fHg(hkl) We have collecting data on the native and derivative crystals. We know the coordinates of Hg. How many unknown quantities remain? SIR Phasing H K L |FP| 9 2 1 486 FP=FP·Hg(+) - fHg(+) |FPH (+)| |FPH (-)| |FPH (+)| |FPH (-)| 586 611 536 499 Imaginary axis 500 |FP | 250 Real axis -500 -250 250 -250 -500 500 SIR Phasing H K L |FP| 9 2 1 486 FP=FP·Hg(+) - fHg(+) |FPH (+)| |FPH (-)| |FPH (+)| |FPH (-)| 586 611 536 499 Imaginary axis 500 |FP | +f’+if[e2pi*(h(x)+k(y)+l(z))+ fHg=fHg e2pi*(h(-x)+k(-y)+l(½+z))+ e2pi*(h(½-y)+k(½+x)+l(¾+z)+ e2pi*(h(½+y)+k(½-x)+l(¼+z)+ e2pi*(h(½-x)+k(½+y)+l(¾-z)+ e2pi*(h(½+x)+k(½-y)+l(¼-z)+ e2pi*(h(y)+k(x)+l(-z)+ e2pi*(h(-y)+k(-x)+l(½-z)] 250 Real axis -500 -250 x,y,z are Hg coordinates from Patterson map (0.197, 0.755, 0.935) f’ and f” are anomalous scattering corrections specific for wavelength used. fHg is a real number proportional to the number of e- in Hg 250 -250 -500 500 SIR Phasing H K L |FP| |FPH (+)| |FPH (-)| 9 2 1 486 586 611 fHg ≠ |FP|-|FPH (+)| FP=FP·Hg(+) - fHg(+) |FPH (+)| |FPH (-)| 536 499 Imaginary axis 500 |FP | No! [e2pi*(h(x)+k(y)+l(z))+ fHg=fHg e2pi*(h(-x)+k(-y)+l(½+z))+ e2pi*(h(½-y)+k(½+x)+l(¾+z)+ e2pi*(h(½+y)+k(½-x)+l(¼+z)+ e2pi*(h(½-x)+k(½+y)+l(¾-z)+ e2pi*(h(½+x)+k(½-y)+l(¼-z)+ e2pi*(h(y)+k(x)+l(-z)+ e2pi*(h(-y)+k(-x)+l(½-z)]+f’+if” 250 Real axis -500 -250 x,y,z are Hg coordinates from Patterson map (0.197, 0.755, 0.935) f’ and f” are anomalous scattering corrections specific for wavelength used. fHg is a real number proportional to the number of e- in Hg 250 -250 -500 500 SIR Phasing H K L |FP| 9 2 1 486 FP=FP·Hg(+) - fHg(+) |FPH (+)| |FPH (-)| |FPH (+)| |FPH (-)| 586 611 536 499 Imaginary axis 500 |FP | |fHg|=282 aHg=58° 250 Real axis 58° fHg(+) -500 -250 250 -250 -500 500 SIR Phasing H K L |FP| 9 2 1 486 FP=FP·Hg(+) - fHg(+) |FPH (+)| |FPH (-)| |FPH (+)| |FPH (-)| 586 611 536 499 Imaginary axis 500 |FP | 250 Real axis -500 -250 250 -fHg(+) -250 Let’s look at the quality of the phasing statistics P·Hg(+)| up to this |F point. -500 500 SIR Phasing H K L |FP| 9 2 1 486 Which of the following graphs best represents the phase probability distribution, P(a)? |FPH (+)| |FPH (-)| |FPH (+)| 586 611 536 4 Imaginary axis 500 |FP | a) 250 0 90 180 270 360 Real axis -500 -250 250 500 -fHg(+) b) -250 0 90 180 270 360 -500 c) |FP·Hg(+)| 0 90 180 270 360 SIR Phasing H K L |FP| 9 2 1 486 The phase probability distribution, P(a) is sometimes shown as being wrapped around the phasing circle. |FPH (+)| |FPH (-)| |FPH (+)| 586 611 536 4 Imaginary axis 500 |FP | 250 0 90 180 270 360 Real axis -500 -250 250 500 -fHg(+) 90 -250 -500 180 0 |FP·Hg(+)| 270 SIR Phasing Which of the following is the best choice of Fp? H K L |FP| 9 2 1 486 |FPH (+)| |FPH (-)| |FPH (+)| 586 611 536 4 Imaginary axis 90 a) 180 500 90 0 |FP | 270 180 -500 90 b) 180 0500Real axis 0 270 270 -500 90 c) 180 0 270 Radius of circle is approximately |Fp| |FP·Hg(+)| SIR Phasing H K L |FP| 9 2 1 486 |FPH (+)| |FPH (-)| |FPH (+)| 586 611 536 4 Imaginary axis 500 |Fp|eia•P(a)da best FP = 90 |FP | a Sum of probability weighted vectors Fp Usually shorter than Fp 180 -500 0500Real axis 270 -500 |FP·Hg(+)| SIR Phasing H K L |FP| 9 2 1 486 |FPH (+)| |FPH (-)| |FPH (+)| 586 611 536 4 Imaginary axis 500 |Fp|eia•P(a)da best FP = 90 a Sum of probability weighted vectors Fp Usually shorter than Fp 180 -500 |Fbest | Real axis 0500 270 -500 |FP·Hg(+)| SIR |Fp|eia•P(a)da best F = 90 a 0 180 270 Best phase Sum of probability weighted vectors Fp SIR Which of the following is the best approximation to the Figure Of Merit (FOM) for this reflection? 90 0 180 a) b) c) d) 1.00 2.00 0.50 -0.10 270 FOM=|Fbest|/|FP| Radius of circle is approximately |Fp| Which phase probability distribution would yield the most desirable Figure of Merit? b) a) 0 90 270 0 180 270 + + 90 180 c) 0 + + 270 180 90 SIR Which of the following is the best approximation to the phasing power for this reflection? Imaginary axis |Fp | Fbest |FPH| Real axis fH |Fp | a) b) c) d) 2.50 1.00 0.50 -0.50 |FPH| Phasing Power = |fH| Lack of closure |fH ( h k l) | = 1.4 Lack of closure = |FPH|-|FP+FH| = 0.5 (at the aP of Fbest) SIR Which of the following is the most desirable phasing power? Imaginary axis |Fp | Fbest |FPH| Real axis fH |Fp | a) b) c) d) 2.50 1.00 0.50 -0.50 |FPH| Phasing Power = |fH| Lack of closure What Phasing Power is sufficient to solve the structure? >1 |FP ( hkl) | = 1.8 |FP•Hg (hkl) | = 2.8 SIR Imaginary axis Which of the following is the RCullis for this reflection? a) b) c) d) |Fp | Fbest |FPH| Real axis fH |Fp | -0.5 0.5 1.30 2.00 |FPH| RCullis = Lack of closure isomorphous difference From previous page, LoC=0.5 Isomorphous difference= |FPH| - |FP| 1.0 = 2.8-1.8 SIRAS Phasing H K L |FP| 9 2 1 486 FP=FP·Hg(-)* - fHg(-)* |FPH (+)| |FPH (-)| |FPH (+)| |FPH (-)| 586 611 536 499 Imaginary axis 500 |FP | |fHg|=282 aHg*=48° 250 Real axis fHg(-)* -500 -250 250 -fHg(+) 48° -250 |FP·Hg(+)| -500 500 SIRAS Phasing H K L |FP| 9 2 1 486 FP=FP·Hg(-)* - fHg(-)* |FPH (+)| |FPH (-)| |FPH (+)| |FPH (-)| 586 611 536 499 Imaginary axis 500 |FP | 250 Real axis -500 -250 -fHg(-)* 250 -fHg(+) -250 |FP·Hg(-)| |FP·Hg(+)| -500 500 SIRAS Isomorphous differences Anomalous differences Which P(a) corresponds to SIR? Which P(a) corresponds to SIRAS? 0 90 180 270 360 Center of inversion ambiguity Remember, because the position of Hg was determined using a Patterson map there is an ambiguity in handedness. The Patterson map has an additional center of symmetry not present in the real crystal. Therefore, both the site x,y,z and -x,-y,-z are equally consistent with Patterson peaks. Handedness can be resolved by calculating both electron density maps and choosing the map which contains structural features of real proteins (Lamino acids, right handed a-helices). If anomalous data is included, then one map will appear significantly better than the other. Patterson map Note: Inversion of the space group symmetry (P43212 →P41212) accompanies inversion of the coordinates (x,y,z→ -x,-y,-z) Choice of origin ambiguity • I want to include the Eu data (derivative 2) in phase calculation. • I can determine the Eu site x,y,z coordinates using a difference Patterson map. • But, how can I guarantee the set of coordinates I obtain are referred to the same origin as Hg (derivative 1)? • Do I have to try all 48 possibilities? Use a Cross difference Fourier to resolve the handedness ambiguity With newly calculated protein phases, aP, a protein electron density map could be calculated. The amplitudes would be |FP|, the phases would be aP. r(xyz)=1/V*S|FP|e-2pi(hx+ky+lz-aP) Answer: If we replace the coefficients with |FPH2-FP|, the result is an electron density map corresponding to this structural feature. r(x)=1/V*S|FPH2-FP|e-2pi(hx-aP) What is the second heavy atom, Alex. When the difference FPH2-FP is taken, the protein contribution is cancelled and we are left with only the contribution from the second heavy atom. This cross difference Fourier will help us in two ways: 1) It will resolve the handedness ambiguity -high peak in difference map calculated with aP in correct hand -only noise in difference map calculated with aP in incorrect hand. 2) It will improve our electron density map of the protein -identify the position of the 2nd heavy atom -include 2 new vector equations for Eu (more accurate aP) Phasing Procedures 1) Calculate phases for site x,y,z of Hg and run cross difference Fourier to find the Eu site. -Note the height of the peak and Eu coordinates. 2) Negate x,y,z of Hg and invert the space group from P43212 to P41212. Calculate a second set of phases and run a second cross difference Fourier to find the Eu site. -Compare the height of the peak with step 1. 3) Chose the handedness which produces the highest peak for Eu. Use the corresponding hand of space group and Hg, and Eu coordinates to make a combined set of phases. MIRAS Phasing H K L |FP| 9 2 1 486 FP=FP·Hg(+) - fHg(+) |FPH (+)| |FPH (-)| |FPH (+)| |FPH (-)| 586 611 536 499 Imaginary axis 500 FP=FP·Hg(-)* - fHg(-)* 250 Real axis -500 fH+f’ f”(-) aH 281 27 53° 250 -250 |FP | = 486 ± 2 H K L 9 2 1 -250 fH+f’ f” aH 100 24 -114° -500 500 SIR SIRAS MIRAS SIR SIRAS MIRAS Density modification A) Solvent flattening. • Calculate an electron density map. • If r<threshold, -> solvent • If r>threshold -> protein • Build a mask • Set density value in solvent region to a constant (low). • Transform flattened map to structure factors • Combine modified phases with original phases. • Iterate MIRAS phased map MIR phased map + Solvent Flattening + Histogram Matching MIRAS phased map MIR phased map + Solvent Flattening + Histogram Matching Density modification B) Histogram matching. • Calculate an electron density map. • Calculate the electron density distribution. It’s a histogram. How many grid points on map have an electron density falling between 0.2 and 0.3 etc? • Compare this histogram with ideal protein electron density map. • Modify electron density to resemble an ideal distribution. Number of times a particular electron density value is observed. Electron density value HOMEWORK Barriers to combining phase information from 2 derivatives 1) Initial Phasing with PCMBS 1) Calculate phases using coordinates you determined. 2) Refine heavy atom coordinates 2) Find Eu site using Cross Difference Fourier map. 1) Easier than Patterson methods. 2) Want to combine PCMBS and Eu to make MIRAS phases. 3) Determine handedness (P43212 or P41212 ?) 1) Repeat calculation above, but in P41212. 2) Compare map features with P43212 map to determine handedness. 4) Combine PCMBS and Eu sites (use correct hand of space group) for improved phases. 5) Density modification (solvent flattening & histogram matching) 1) Improves Phases 6) View electron density map