Neural Network Analysis of Flow Cytometry Immunophenotype Data
advertisement

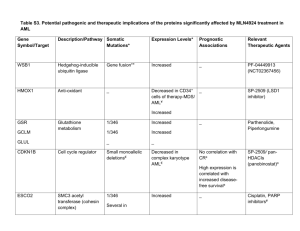
IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING, VOL. 43, NO. 8, AUGUST 1996 Neural Network Analysis of Flow Cytometry Immunophenotype Data Ravi Kothari,* Member, IEEE, Hernani Cualing, and Thiagarajan Balachander Mehrshad Mokhtaran M.D. Acute Leukemia • Definition – Malignant Event – Replace the bone marrow with blast – Clinical Complication: Anemia, Infection, Bleeding – Rapidly fatal – With appropriate therapy, the natural history can be markedly altered, and many patient can be cured. Acute Leukemia • Etiology: – Radiation – Oncogenic Viruses – Genetic and Congenital Factors – Chemical and Drugs Acute Leukemia • Incidence: – – – – – – – – – Annual new case (All Leukemia): 8 to 10 per 100,000. Remained static over the past three decades. ALL:11% CLL:29% AML:46% CML:14% 3% of all cancer in United States ALL is most common cancer in children(<15y) ALL is second cause of death in children(<15y) ALL has tow maximum incidence per age AML gradually increases with age Half of AML cases occur in patients younger than 50 y Acute Leukemia • Pathophysiology: Acute Leukemia • Classification – Morphology – Cytochemistry – Cell-surface markers – Cytoplasmic markers – Cytogenetics – Oncogene expression Acute Leukemia • Must important Distinction is between: AML & ALL – Clinical behavior, prognosis, response to therapy • AML (FAB) – – – – M0, M1, M2, M3: Increasing degree of differentiation M4, M5: Monocytic lineage M6: Erytroid cell linage M7: Acute Megakaryocytic Leukemia • ALL (FAB) – L1 – L2 – L3 Acute Leukemia • Cell-surface Markers: – AML • Normal immature myeloid cells and blast cells from most patient with AML: CD13, CD14, CD33, CD34 • M6, M7: Antigens restricted to red cell and platelet lineage • AML may express: HLA-DR antigen • 10-20%: B- or T-cell lineage – ALL • 60% of ALL: CALLA(CD10) (early pre-B-cell differentiation state) • Pre-B-cell ALL: 20% CALLA-positive that have intracytoplasmic immunoglobulin • B-cell ALL(5%): Immunoglobulin on cell surface • T-cell ALL(20%): CD5, CD3 or CD2 (normal early T-cell) • Null cell ALL (15%): Fail to express CALLA, B- , T-cell markers • 25% of ALL: Myeloid antigens Acute Leukemia • Cytogenetics and Molecular biology: Acute Leukemia • Clinical Manifestations: – Decreaseing normal marrow function: • Anemia: Fatigue, pallor, headache, angina or heart failure • Thrombocytopenia: Bleeding(petechiae, ecchymosess, bleeding gums, epistaxis) • Granulocytopenic(AML>ALL) : Infections (Bacterial) – Invasioning of normal organs by leukemic blasts (ALL>AML): • • • • • • • Enlargement of lymph nodes, liver, spleen Bone pain Skin (Leukemia cutis) Leukemic meningitis: Headache, nausea CNS (particular in relapse): palsies and seizures Testicular involvement (particular in relapse) Any soft tissue (AML>ALL): Chloroma, myeloblastoma – Specific subtype of leukemia: • M3: DIC (Disseminated intravascular coagulation) Acute Leukemia • Laboratory Manifestations: – CBC – Bone marrow aspiration and biopsy – PT (Prothrombin Time) & PTT (Partial Thromboplastin Time) – LDH (Lactate dehydrogenase) –… Acute Leukemia • Treatment: – Combination Chemotherapy – Bone Marrow Transplantation – Stabilization: • Hematological • Metabolical • Psychological • • • • • • Introduction Data Collection Classifier Design Results Discussion Conclusion Introduction • • • • Immunophenotype data Flow cytometry Lineage & Differentiation ALL: Immature (CALLA+), Pre-B, Mature-B, T-Lymphoblastic • Response to chemotherapy • AML: M1,M2,…,M8 • No relevant prognosis Data Collection • Flow cytometry immunophenotype data of cases with leukemia or reactive bone marrow were collected retrospectively from computerized archival database. • Selection Criterion: – Confirmed diagnosis – Complete flow cytometry antibody panel result • Total cases: 170 – 151 leukemia and 19 nonleukemia – 62 children and 89 adults – 81 males and 70 females First Phase • Lineage Categories • Categorize into: – Reactive – ALL – Remission – Mixed AML-ALL – AML Second Phase • Categorize the ALL Cases into subcategories based on differentiation • Categorize into: – Pre-B – CALLA+ – T Phenotype • Not include: Mature-B (Difficulty in obtaining sufficient data for meaningful interpretation) Data • • • • Validation / Training set size = 33-50% Only Bone marrow phenotypes (Most Sensetive specific) Excluded: Peripheral blood and cerebro-spinal fluids immunophenotype Flow cytometry immunophenotype data: – Mean fluorescence intensity of a minimum of 10000 cells analyzed using either a red or green fluorescence tagged antibody Data • 27 Standardized and most commonly used monoclonal antibodies with defined specificities. • Not all of these are utilized for each case. • Average of 15 antibodies for each case. • At least ten antibodies are commonly used for acute leukemia as a standard practice. • With a zero value if an antibody was not used • An additional binary input denoting past diagnosis of leukemia, were used as input a neural network classifier. Classifier Design • A feed-forward neural network • Trained using back propagation algorithm Classifier • How many hidden layer neurons are needed for a particular task? – Having a large number of redundant weights leads to over fitting Classifier • Given a network with a certain number of inputs, hidden layer neurons, and output, needed how many training sample are to achieve good generalization? • For accuracy of (1-ε): p ≥ O(W/ε) p: Number of training sample. W: Total number of weights in the network. Classifier • Perturbation: To generate a large number of cases by introducing small variation in actual cases. • Optimal Brain Damage: The weight which least increase the error can be eliminated • Optimal Brain Surgeon: The sensitivity of an interconnection is expressed as the cumulative sum of the changes experienced by a weight, during training. • Weight Decay: Each weight has a tendency to decay to zero with a rate proportional to the magnitude of the weight. Classifier • Inputs: 27 + 1 • Hidden: 50 Progressively increasing the number of hidden neurons until acceptable performance was achieved on training data. • Output: – First phase (Based on lineage): 5 – Second phase (Based on differentiation): 3 • Learning rate (η): 0.1 • Weight Decay Coefficient (λ): 0.05 Results • Mean error was acceptably low (0.0001) in both the cases. • First phase weights : – Total: 1650 – Nonzero: 1106 – Very small value(<0.1): 544 • Second phase weights : – Total: 1550 – Nonzero: 446 – Very small value(<0.1): 1104 Fig. 2. Performance of the network for categorization into reactive and the lineage categories of leukemia (ALL, Remission, Mixed AML-ALL, and AML). Fig. 3. Performance of the network for categorization of ALL cases into subcategories based on differentiation (Pre-B, CALLA+, and T Phenotype). Result • Generalization Error: – First phase: 10.3% – Second phase: 10.0% • Back propagation without the complexity regulation term (Weight Decay): – Generalization performance was poor Discussion • Clustering-based methods fall into one of two categories: – Partitioning – Hierarchical Discussion • Partitioning: – e.g., k-means, c-means fuzzy clustering – Divide the inputs, so that members of a cluster are close to each other and far away from other clusters – The shared specificity of some monoclonal antibodies make this extremely difficult. Discussion • Hierarchical: – e.g., centroid sorting, linkage methods – Try to merge two closest data points together at each step, and repeat the process until there is only one cluster. – Have a better chance of succeeding due to the variability in immunophenotype data – An error in merging made earlier on is propagated throughout. Conclusion • Off line retraining • Extract rules from trained networks